SPR (Biacore)
SPR (Biacore) Assays: Label-Free Pharmacology for Lead Characterisation and Candidate Selection.
This label-free technique measures the real-time binding affinity of drug leads and candidates using Surface Plasmon Resonance (SPR).
SPR (Biacore) can be used with biological and small molecule therapeutics. Binding parameters can help characterise drug leads to select higher affinity compounds for further chemistry exploration, or guide selection of drug candidates with the slowest dissociation rates to ensure maximal target engagement.
BioLayer Interferometry (BLI) provides an alternative label-free technique. It has more assay design flexibility than SPR but has more limited options for protein immobilisation onto the sensor surface.
Our Areas of Expertise:
- Antibodies, fAbs and ScFv fragments
- Viral antigen and antibody detection
- Cytokines and receptor tyrosine kinases (RTKs) – small molecules and biologics
- Chromatin modification enzymes (epigenetics) – small molecules
- Transcription factors & nuclear hormone receptors – small molecules
- Protein complex formation and disruption – small molecules and biologics
- Plasma biomarker and in vitro diagnostic assays
Assay Formats:
- Direct binding affinities (kinetic: ka, kd, KD; equilibrium: KD)
- Solution phase binding inhibition (IC50 and Ki)
- Concentration analysis and analyte quantitation in biological samples
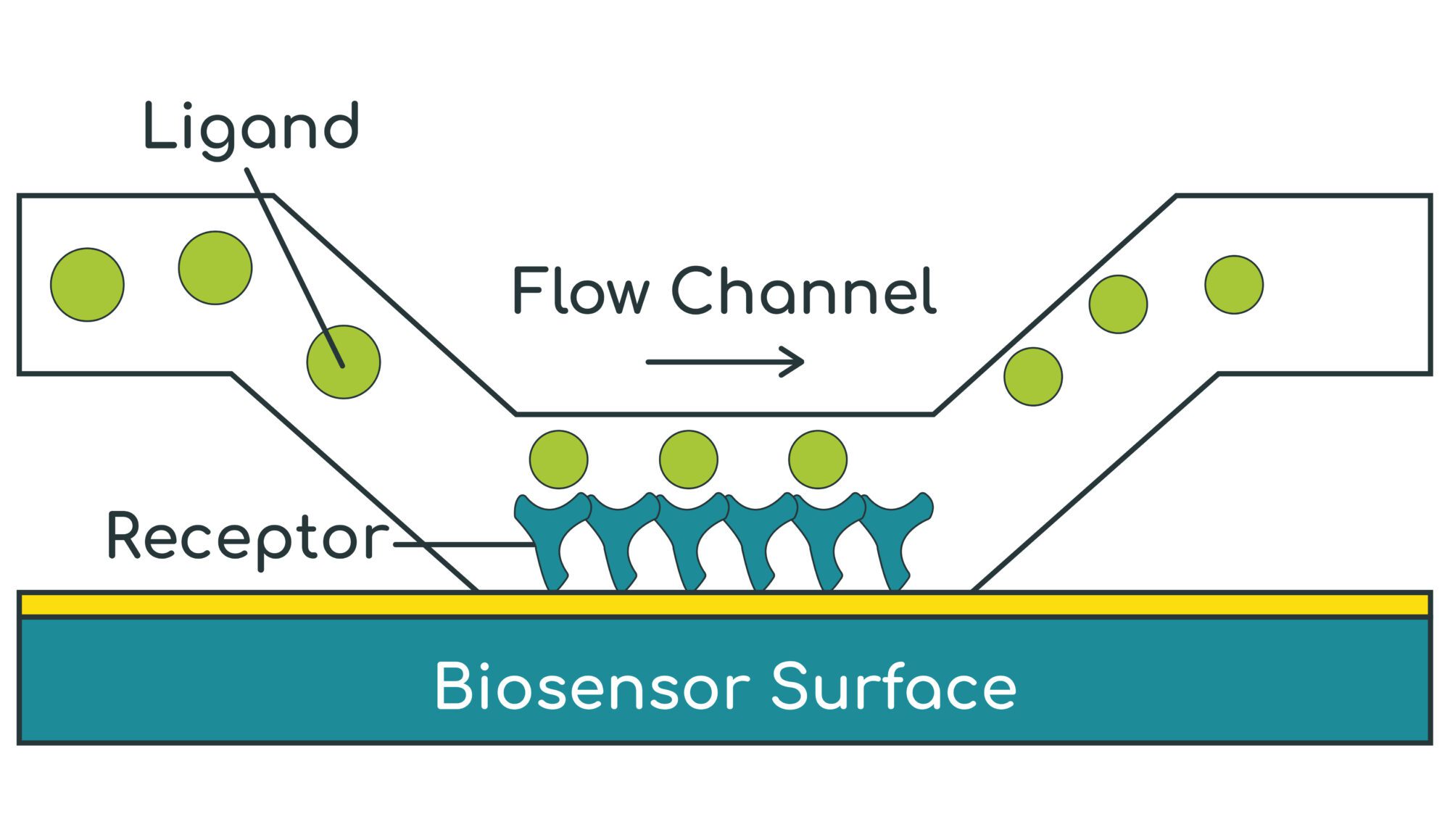
- SPR (Biacore) measures the refractive index of the material within a ~100 nm layer immediately above the gold-coated biosensor surface
- A target protein (receptor) is first immobilised / captured onto the biosensor surface
- A test substance (ligand) is then flowed over the surface to bind to the receptor
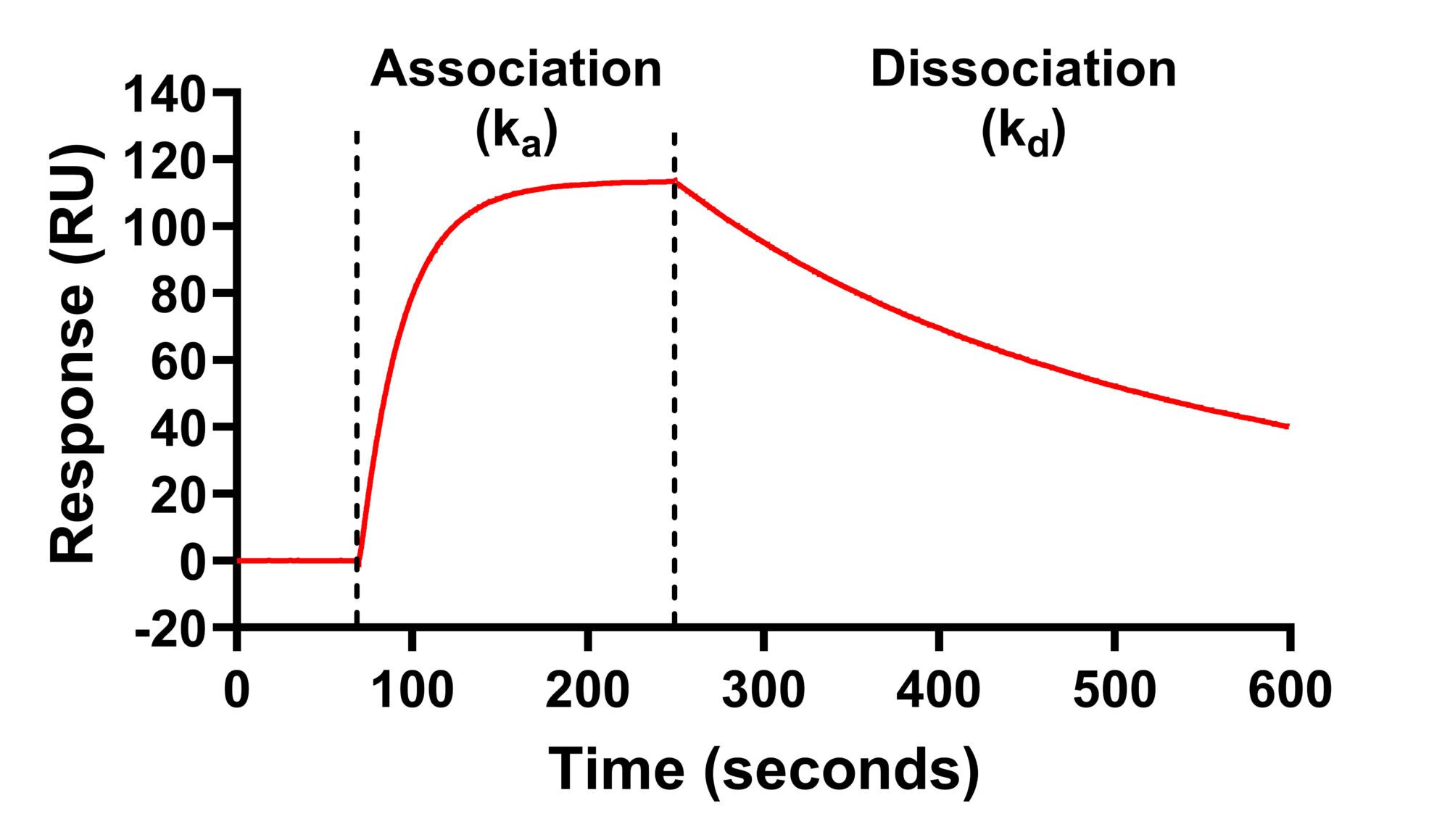
- As the ligand binds to the receptor, the refractive index increases in real-time – this allows the binding association rate (ka) to be measured
- The ligand is then washed off the receptor with buffer and the refractive index falls in real time – this allows the dissociation rate (kd) to be measured
- The kinetic binding affinity constant (KD) is then calculated as kd / ka
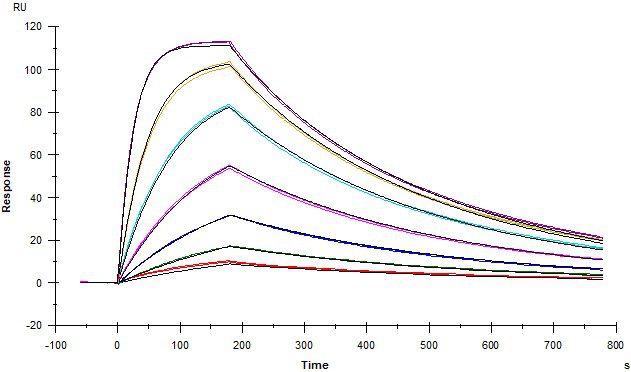
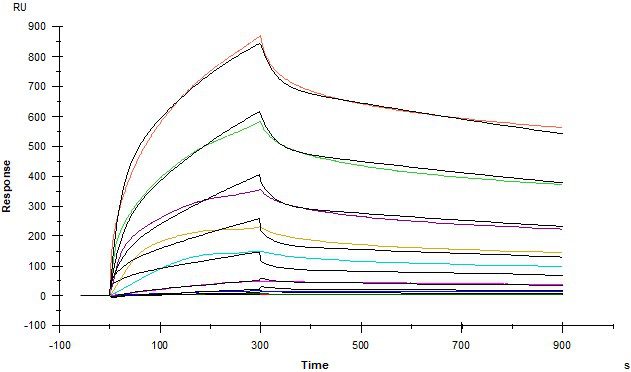
Example Data
Protein – Protein Interactions
Kinetic affinity (KD) and solution-phase competition (IC50 and Ki) analysis
ACE2 and Spike RBD
Angiotensin converting enzyme 2 (ACE2) covalently coupled onto a CM5 sensor. SARS-CoV2 Spike RBD protein binds to ACE2 with a kinetic affinity KD of 30 nM.
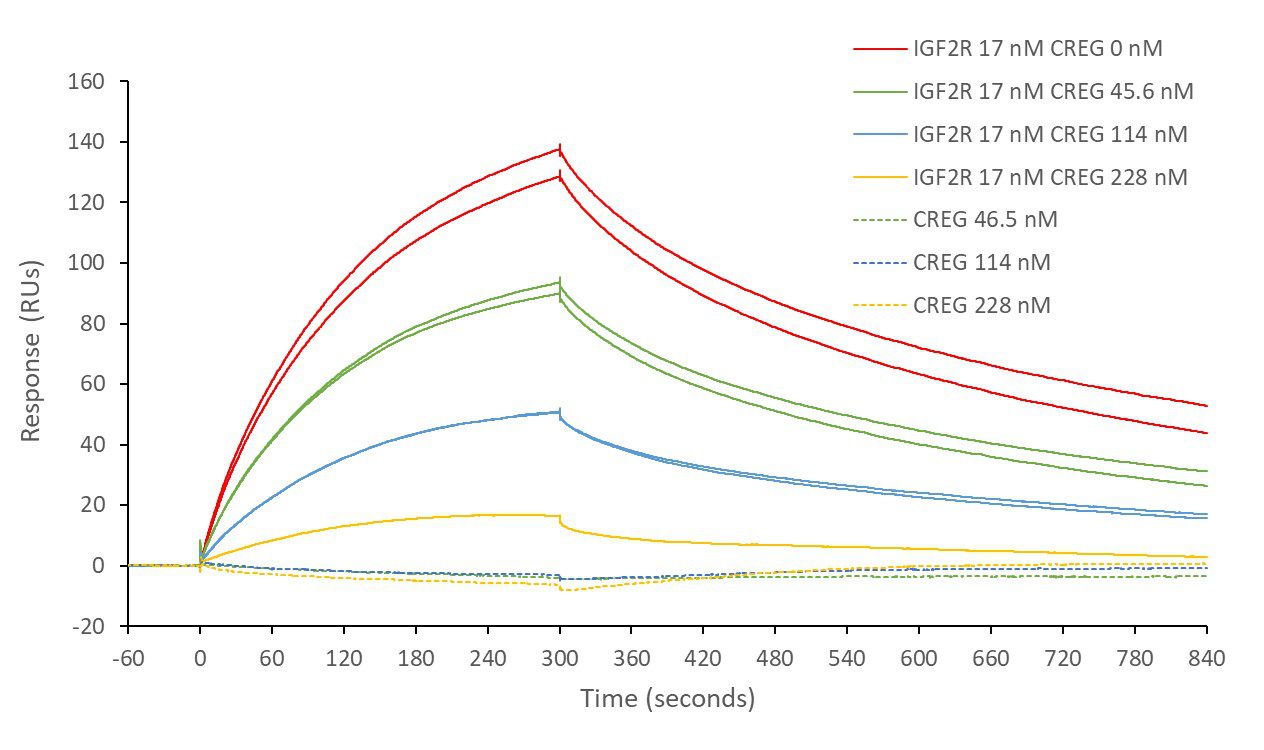
IGF2R and STC1
Insulin-like growth factor 2 receptor (IGF2R) covalently- immobilised onto a CM5 sensor. Stanniocalcin 1 (STC1) binds to IGF2R with a kinetic affinity KD of 16.8 nM.
Antibody – Antigen Interactions (Quantitation in Biological Samples)
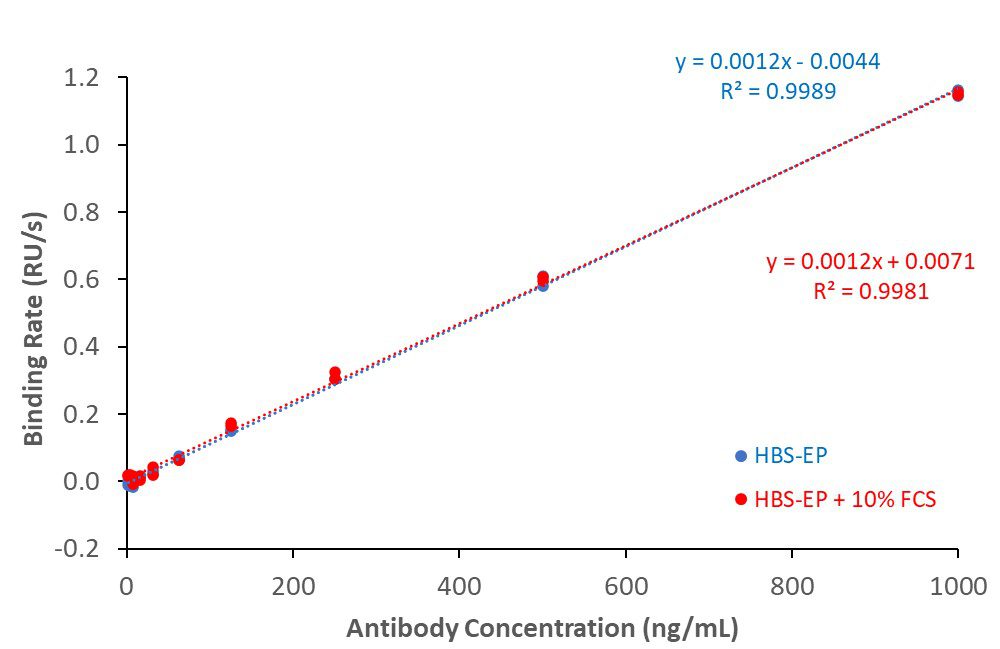
SARS-CoV2 spike RBD – anti-spike antibody binding (in buffer and 10% serum)
His-tagged SARS-CoV2 spike receptor binding domain (RBD) protein covalently coupled to CM5 sensor surface. Rabbit anti-spike binding responses in buffer (HBS-EP) or buffer + 10% serum generated antibody calibration curves. Lower limit of detection (LLD): 3.9 ng/mL; lower limit of quantitation (LLQ): 15.6 ng/mL.
Receptor – Ligand Interactions
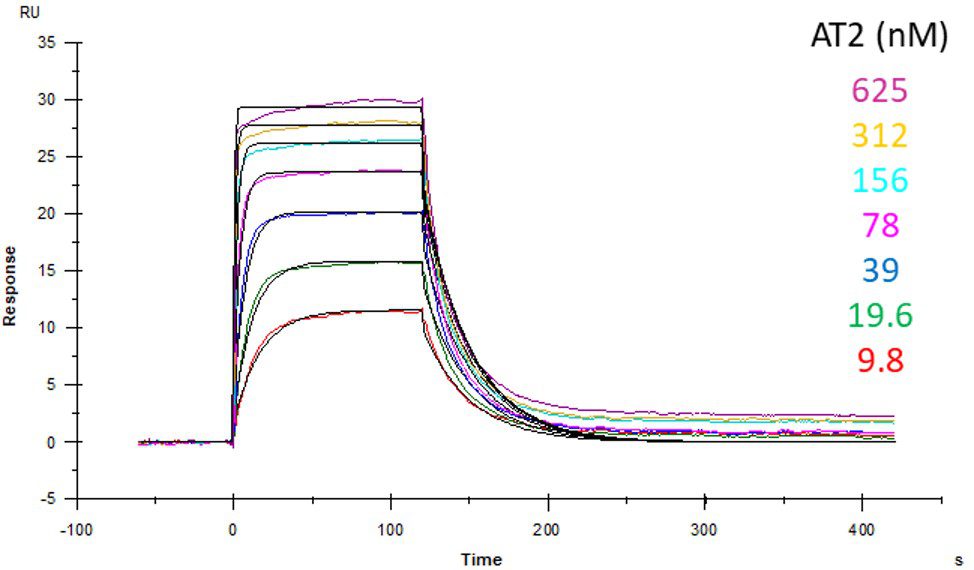
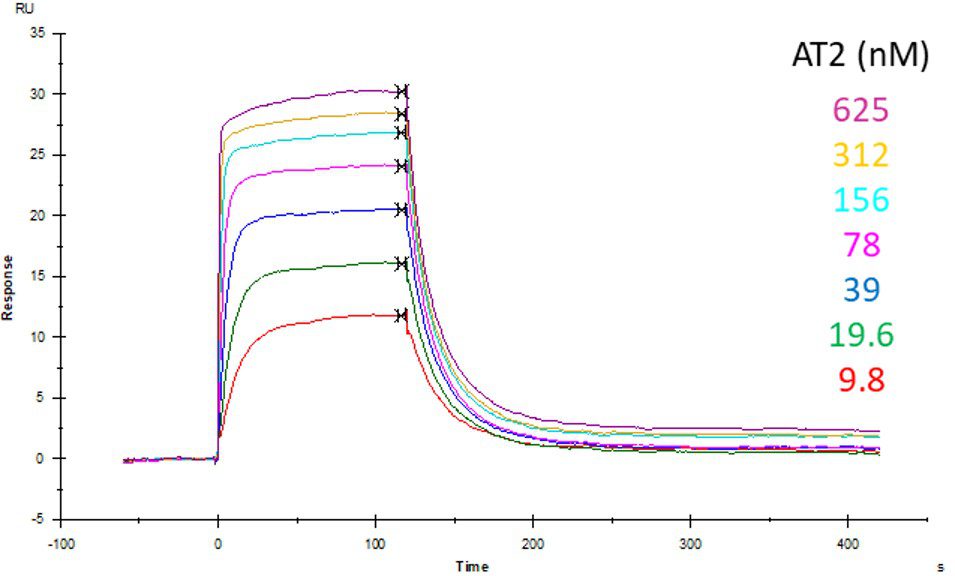
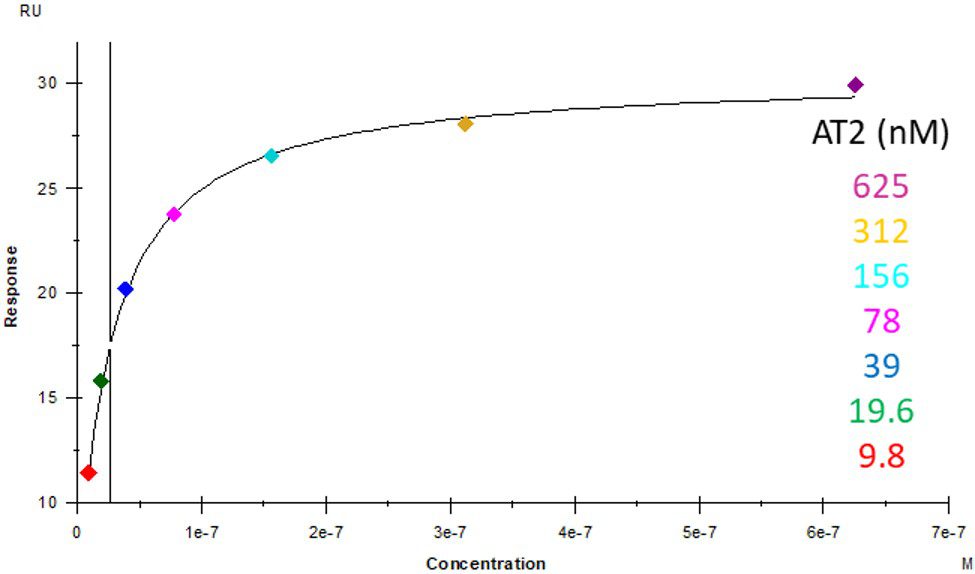
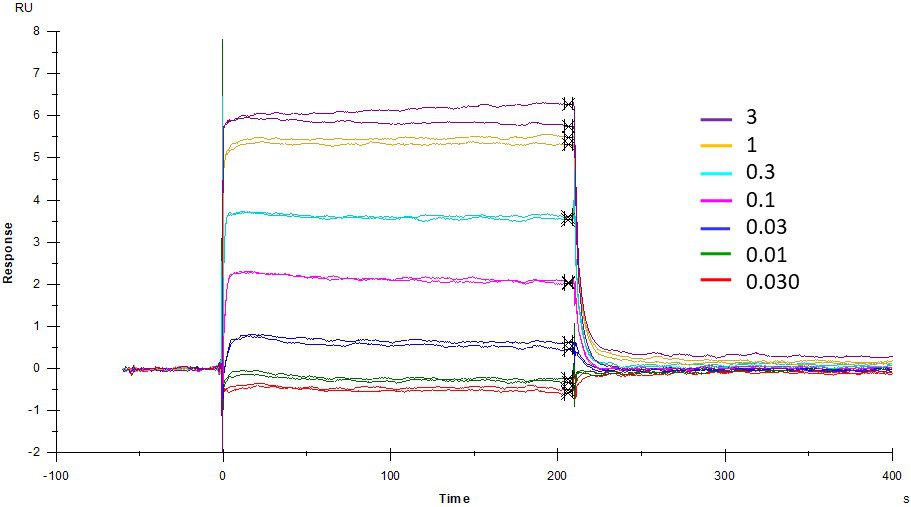
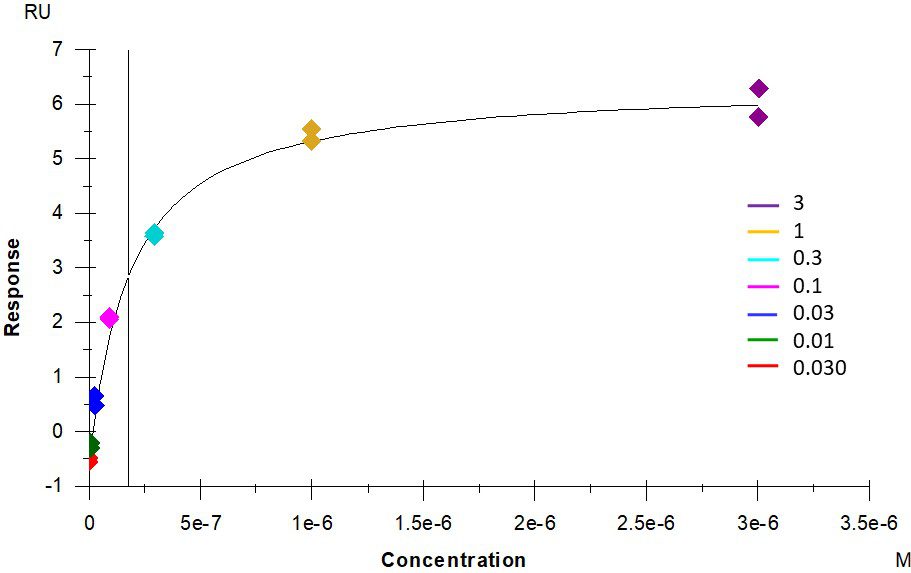
ACE2 + AT2 – kinetic affinity and equilibrium binding analysis
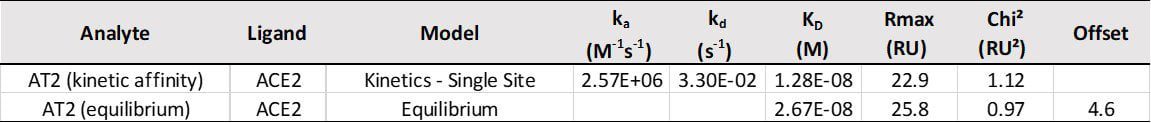
ACE2 covalently immobilised onto a CM5 sensor. Angiotensin II (AT2; 1046.2 Da) binding responses to ACE2 were fitted to a single-site kinetic affinity model (left) to give a KD of 12.8 nM. Fitting the same data to an equilibrium binding model (centre) using the response amplitudes marked at ‘x’, plotted on a linear saturation binding model (right), gives a KD (concentration at half-maximal response) of 26.7 nM.
Receptor – Small Molecule Interactions
Protac ligand binding to Von Hippel–Lindau (VHL) protein
Protac ligand MZ1 (1002.64 Da) binding to covalently-immobilised Von Hippel-Lindau (VHL) protein gives a saturable binding response of 7.0 RUs with a KD of 178 nM. In contrast, the non-binding control, cis-MZ1 (1002.64 Da) gives a non-saturable, non-specific binding response with an estimated KD > 240 µM.