Radioligand Binding Assay
Radioligand binding assays are considered the gold standard for measuring the affinity of ligand binding to a target receptor, due to their robustness and sensitivity. There are three types of radioligand binding assay: competitive, saturation and kinetic.
Competitive radioligand binding assays are used to determine the relative affinities (Ki values) of test compounds for binding to a receptor site in a membrane homogenate or cells. They are performed by incubating a range of concentrations of the unlabeled test compound with a fixed concentration of radioligand. Then measuring the IC50 (nM) of the test compound to competitively inhibit binding of the radiolabeled ligand to its receptor. Our standard study design is for ten concentrations of the unlabeled test article over a five-log unit range, duplicate determinations.
Saturation radioligand binding assays allow determination of the number of binding sites, Bmax (fmol/mg protein or sites per cell), in the tissue or cultured cells. Also the dissociation constant, Kd (nM), of the radioligand, to measure affinity. They are performed with increasing concentrations of the radioligand to directly measure the level of receptor binding. Our standard study design is for eight concentrations of the labeled test article over a two-log unit range, duplicate determinations.
Kinetic radioligand binding assays are used to determine the association and dissociation rates of a radiolabeled ligand from a receptor, to provide additional information for optimizing assay conditions. Our standard study design is for eight to twelve time points to obtain association or dissociation rate constants, duplicate determinations.
There are two main variations in protocol: filtration assay or scintillation proximity assay
Filtration assay protocol is to incubate the membrane homogenate or cultured cells with both the radioligand and the competing test compound (if any). Once equilibrium has been attained, receptor-bound radioligand is separated from free (unbound) radioligand using a 96-well filtration apparatus. The receptor-bound radioactivity trapped on the filters is counted and the results plotted to obtain IC50 and Ki values for the test compound, or to determine Bmax for the receptor levels. Filtration assays work well for membrane-bound receptors since membrane microsomes in the homogenate are readily trapped on the glass fiber filters.
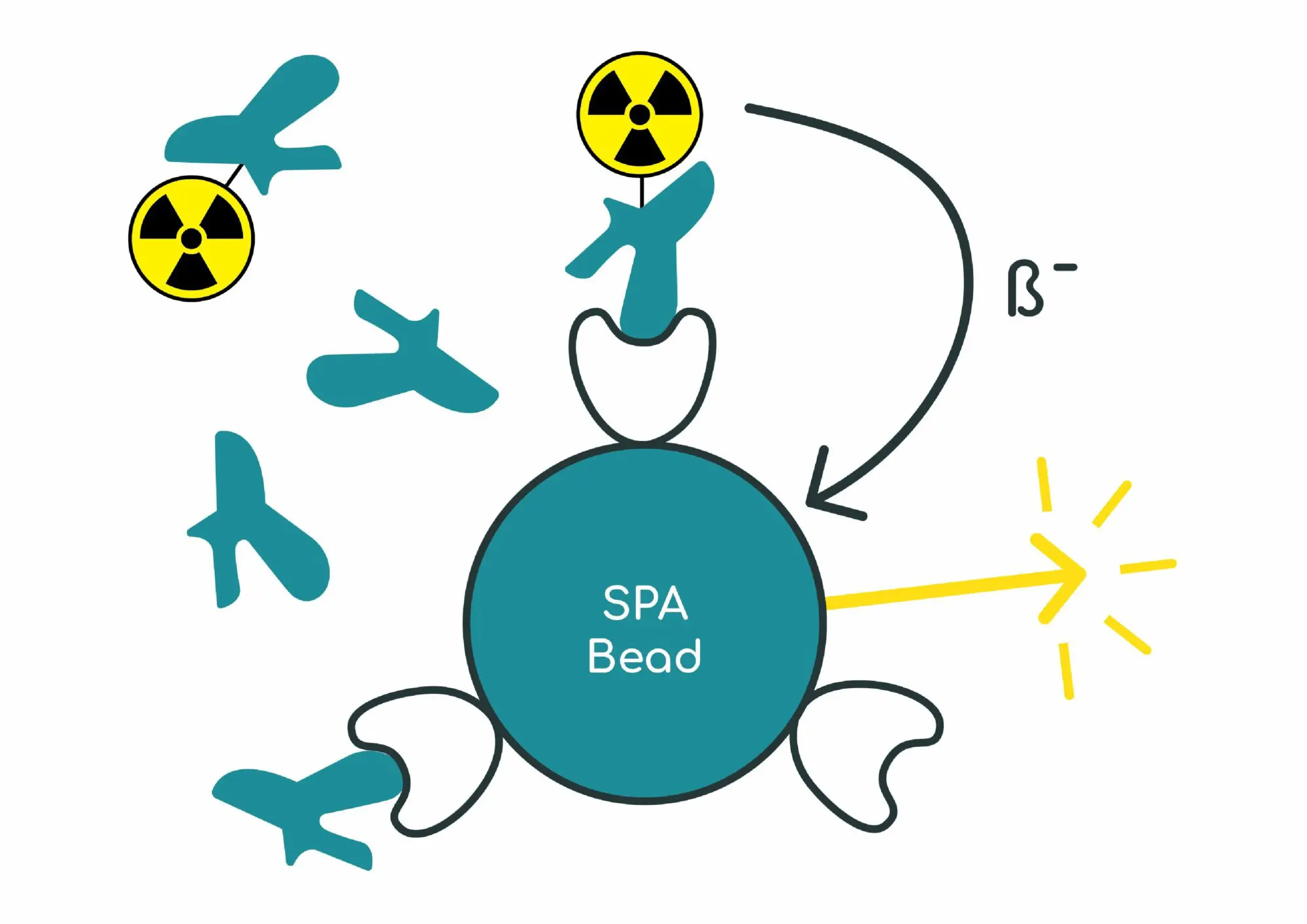
Scintillation proximity assay protocol does not require separating bound from free radioligand. The enzyme, receptor or cultured cells are absorbed or bound onto the surface of scintillation proximity assay (SPA) beads. The receptor-coated beads are then incubated together with a radioligand and the competing test compound. After equilibrium has been reached, light emission from radioligand bound to a receptor on the surface of the beads is counted using a 96 well plate-reader. SPA assays can be applied both to soluble cytoplasmic receptors and membrane-bound receptors, depending on the method used for immobilization onto the bead.
Example Data: Competitive Radioligand Binding Assay
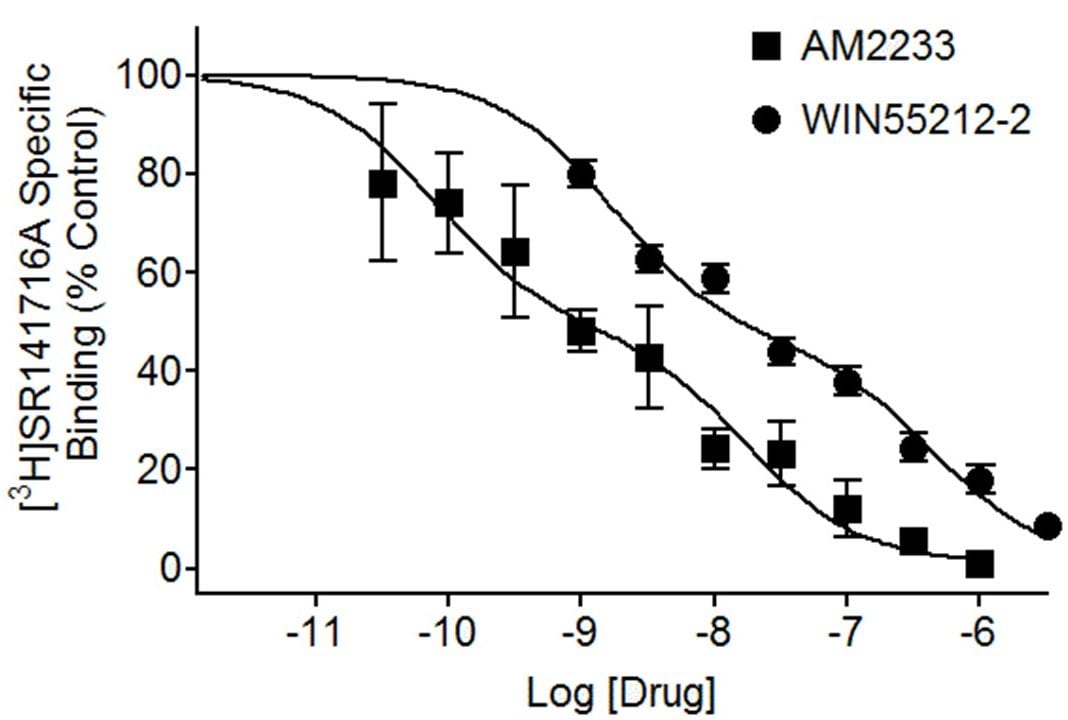
Displacement of [3H]SR141716A binding to CB1 cannabinoid receptors in rat brain homogenate by the cannabinoid agonists AM2233 and WIN55212-2. Displacement curves are shown fitted to a two-site model with Ki values of 0.08 nM and 17 nM for AM2233 and 2 nM and 43 nM for WIN55212-2.
Example Data: Saturation Radioligand Binding Assay
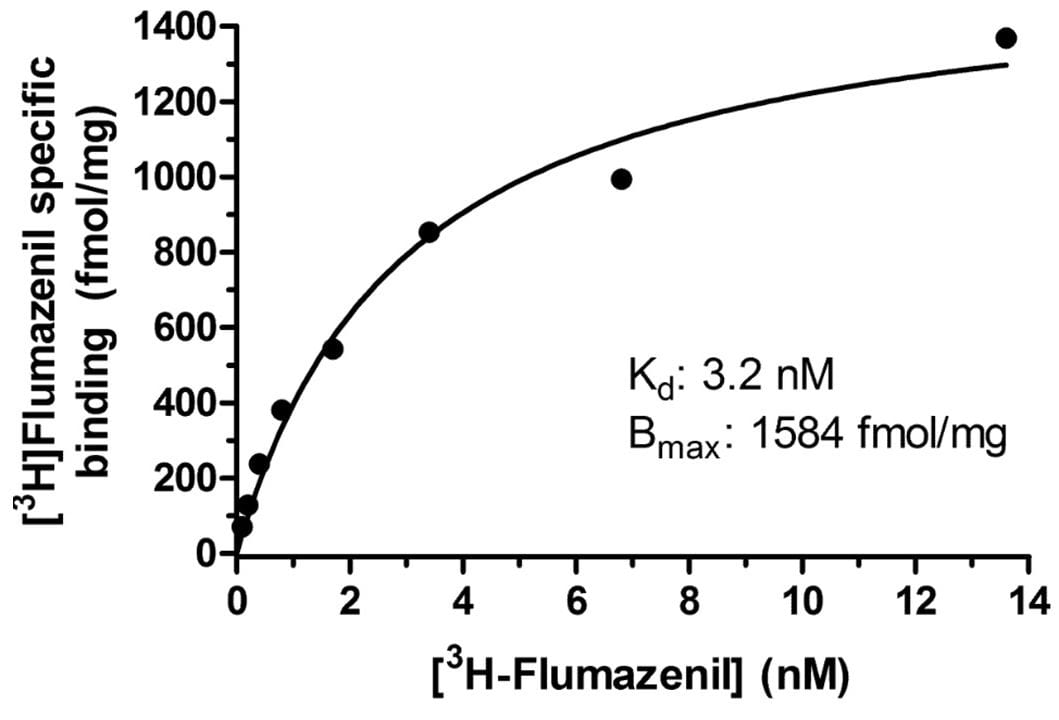
Specific binding of a radioligand for the brain BZD receptor in human post-mortem brain tissue. Binding was to a single saturable site with Kd 3.2 nM and Bmax 1584 fmol/mg protein.
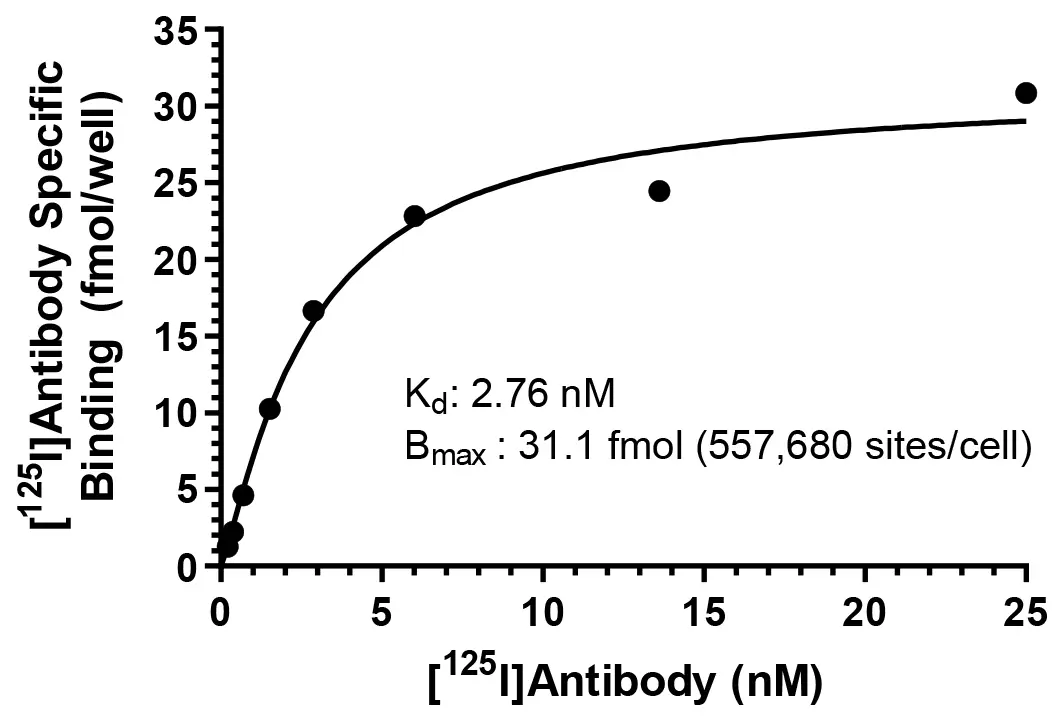
Specific binding of a [125I]-labeled monoclonal antibody to cell surface antigen sites on SKBR3 cells.
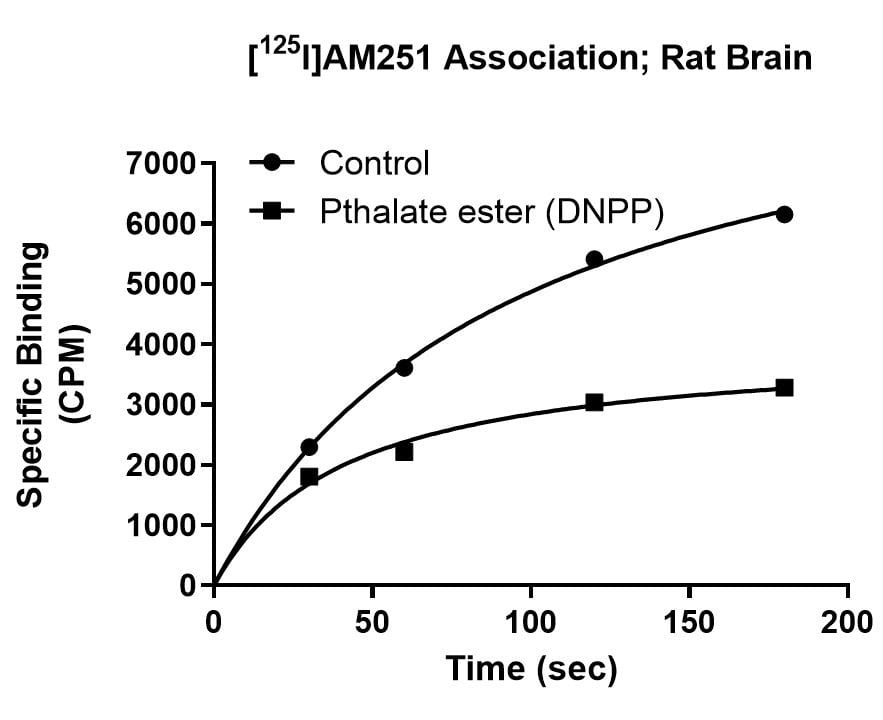
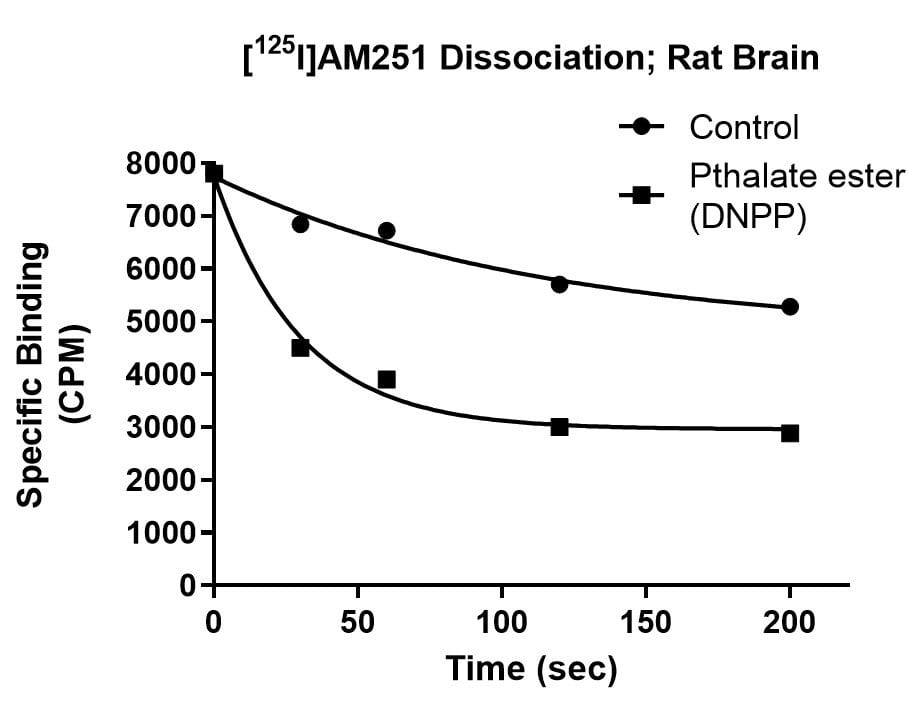
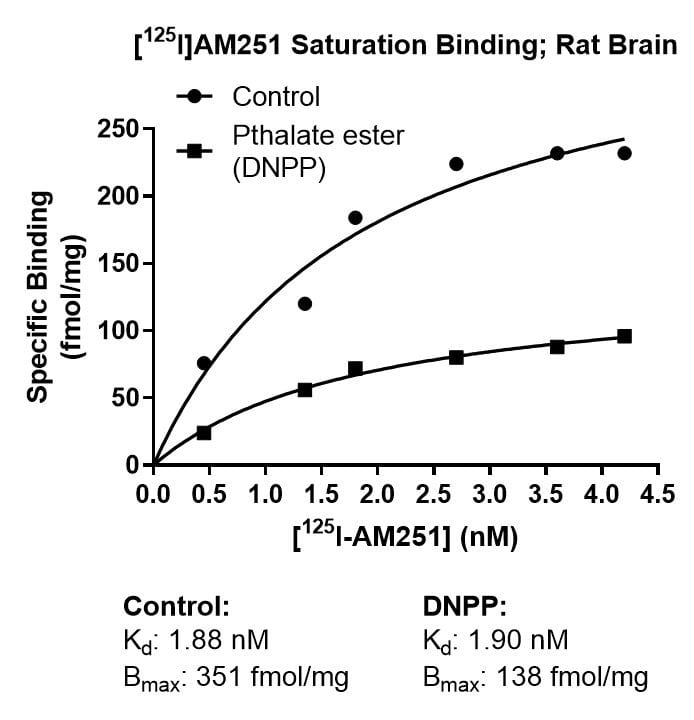
Kinetics Example
Effect of Di-n-pentyl phthalate (DNPP; 40 µM) on association rate, dissociation rate and saturation binding of the cannabinoid ligand [125I]AM251 in rat brain. The enhanced dissociation rate and lowered Bmax for [125I]AM251 binding in the presence of the inhibitor is consistent with an allosteric binding site for DNPP on the CB1 receptor.