Angiotensin II and SARS-CoV-2 ‘spike’ protein bind to independent sites on angiotensin converting enzyme 2.
We used the Biacore Surface Plasmon Resonance (SPR) technique to show that the peptide angiotensin II (AT2, that is normally truncated by the ACE2 receptor) and COVID-19 / SARS-CoV-2 spike protein bind to different sites on the enzyme. When pre-mixed, both AT2 and spike protein are still able to bind to ACE2 without blocking each other. Furthermore, if spike is bound to ACE2 first, and AT2 is then added while spike it still bound, spike does not hinder AT2 binding. This indicates that AT2 and spike protein are likely to bind to non-competiting sites on ACE2. Consequently, small, drug-like molecules binding to the AT2 site on ACE2 are unlikely to be effective as antiviral agents to prevent ‘spike’ binding.
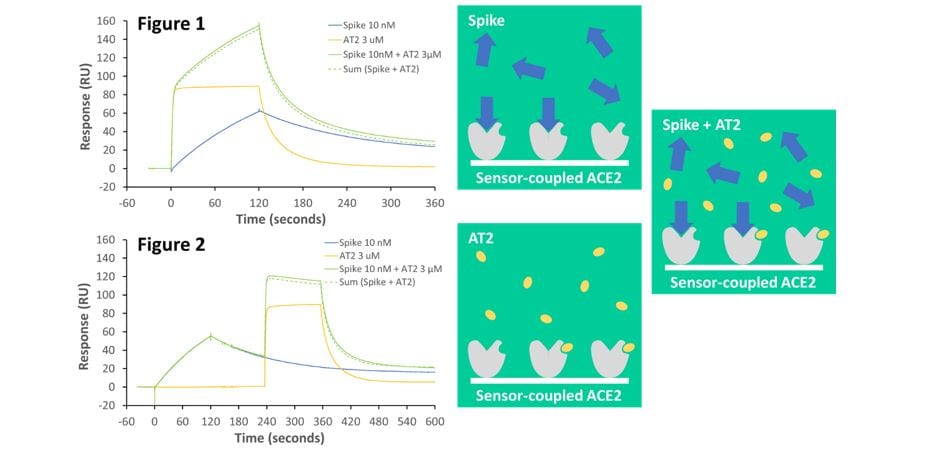
Figure 1. Both angiotensin II (AT2) (3 µM; yellow line) and spike protein (10 nM; blue line) individually bind to immobilised ACE2. However, pre-mixing and co-injecting spike protein and AT2 gives a response (solid green line) that is equivalent to the sum of both individual responses (dotted green line). This demonstrates that spike and AT2 bind independently.
Figure 2. Even when spike protein is bound to ACE2 first, and remains bound when AT2 is added subsequently, AT2 binding is not blocked by bound spike protein. This shows that the independent spike and AT2 binding sites do not overlap.